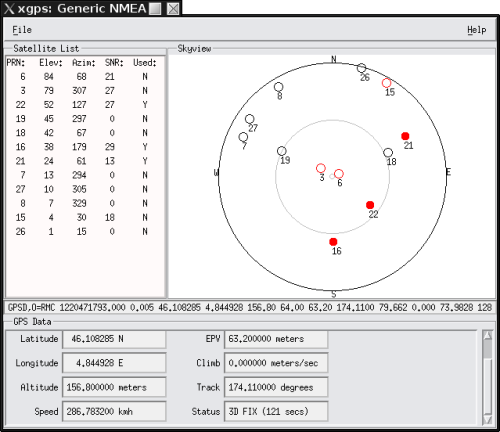
Direct access to tty
By default your gps unit is assigned to /dev/ttyUSB0, but you should check to make sure:
>dmesg | grep -i USB
[11250.384023] usb 4-2: new full-speed USB device number 2 using uhci_hcd
[11250.669968] usbcore: registered new interface driver usbserial
[11250.669991] USB Serial support registered for generic
[11250.670030] usbcore: registered new interface driver usbserial_generic
[11250.670033] usbserial: USB Serial Driver core
[11250.673280] USB Serial support registered for pl2303
[11250.685151] usb 4-2: pl2303 converter now attached to ttyUSB0
[11250.685174] usbcore: registered new interface driver pl2303
[11250.685177] pl2303: Prolific PL2303 USB to serial adaptor driver
Set access, change the baud rate and put the GPS in NMEA mode. Look at some of the output:
>sudo chmod 666 /dev/ttyUSB0 && stty -F /dev/ttyUSB0 ispeed 4800
>cat /dev/ttyUSB0
$GPGSA,A,3,15,14,09,18,21,24,22,06,19,,,,1.9,1.2,1.4*36
$GPGSV,3,1,12,18,71,317,34,24,63,112,31,21,52,212,28,15,42,058,34*71
$GPGSV,3,2,12,22,37,301,28,14,21,250,24,09,07,035,21,19,06,316,13*73
$GPGSV,3,3,12,06,05,279,18,26,01,060,,03,01,294,,28,01,024,*7F
$GPRMC,224355.000,A,1825.0332,N,02111.3093,W,0.13,134.34,050513,,,A*75
$GPGGA,224532.000,1825.0301,N,02111.3093,W,1,08,1.3,39.8,M,-73.7,M,,0000*25
$GPGSA,A,3,15,14,09,18,21,24,22,06,19,,,,1.9,1.2,1.4*36
$GPRMC,224356.000,A,4225.0330,N,02111.3093,W,0.33,145.51,050513,,,A*73
$GPGGA,224357.000,1825.0328,N,07111.3092,W,1,09,1.2,81.8,M,-33.7,M,,0000*5F
$GPGSA,A,3,15,14,09,18,21,24,22,06,19,,,,1.9,1.2,1.4*36
$GPRMC,224357.000,A,1825.0328,N,02111.3092,W,0.28,151.99,050513,,,A*71
Use Ctrl-C to stop the process.
Pick a sentence format that contains the coordinate information you wish to extract. I chose GPGGA.
>cat /dev/ttyUSB0 | grep GPGGA
$GPGGA,124532.000,1825.0301,N,02111.3093,W,1,08,1.3,39.8,M,-73.7,M,,0000*25
$GPGGA,124532.000,1825.0301,N,02111.3093,W,1,08,1.3,39.8,M,-73.7,M,,0000*25
$GPGGA,124532.000,1825.0301,N,02111.3093,W,1,08,1.3,39.8,M,-73.7,M,,0000*25
$GPGGA,124532.000,1825.0301,N,02111.3093,W,1,08,1.3,39.8,M,-73.7,M,,0000*25
$GPGGA,124532.000,1825.0301,N,02111.3093,W,1,08,1.3,39.8,M,-73.7,M,,0000*25
$GPGGA,124532.000,1825.0301,N,02111.3093,W,1,08,1.3,39.8,M,-73.7,M,,0000*25
$GPGGA,124532.000,1825.0301,N,02111.3093,W,1,08,1.3,39.8,M,-73.7,M,,0000*25
$GPGGA,124532.000,1825.0301,N,02111.3093,W,1,08,1.3,39.8,M,-73.7,M,,0000*25
$GPGGA,124532.000,1825.0301,N,02111.3093,W,1,08,1.3,39.8,M,-73.7,M,,0000*25
$GPGGA,124532.000,1825.0301,N,02111.3093,W,1,08,1.3,39.8,M,-73.7,M,,0000*25
You can extract to a text file using:
cat /dev/ttyUSB0 | grep --line-buffered GPGGA > output.txt
Now with further munging you can extract the coordinates. I’ll save that for the next section where I use gpsd.